Advanced usage example
This example illustrates how to creating a custom disease module. It is a translation of Starsim’s Diseases developer tutorial to R.
To recap that tutorial, the main customizations we are performing here are:
- In
__init__()we added the extra pars and states needed for our model. - We defined infectious to include both infected and exposed people - this means that we can just reuse the existing logic for how the SIR model handles transmission.
- We updated
step_state()andstep_die()to include changes to the exposed state. - We rewrote
set_prognoses()to include the new exposed state.
library(starsim)
load_starsim()
# Create a new class for an SEIR disease
SEIR <- PyClass("SEIR",
inherit = ss$SIR, # Inherit from the SIR class
defs = list(`__module__` = "SEIR",
# Define the __init__ method
`__init__` = function(self, pars = py_none(), ...) {
ss$SIR$`__init__`(self) # Call the parent init method
# Parameters specific to SEIR: duration of exposure
self$define_pars(
dur_exp = ss$lognorm_ex(0.5), # Define an explicit ("ex") lognormal distribution with a mean of 0.5
)
self$update_pars(pars, ...)
# Additional states beyond the SIR ones
self$define_states(
ss$State('exposed', label='Exposed'),
ss$FloatArr('ti_exposed', label='TIme of exposure'),
)
return(py_none())
},
# Make all the updates from the SEIR model
step_state = function(self) {
ss$SIR$step_state(self) # Perform SIR updates
# Additional updates: progress exposed -> infected
infected <- self$exposed & (self$ti_infected <= self$ti) # Check who is scheduled to become infected
self$exposed[infected] = FALSE
self$infected[infected] = TRUE
return(py_none())
},
# Ensure that exposed is set to False for people who die
step_die = function(self, uids) {
ss$SIR$step_die(self, uids) # Perform SIR updates
self$exposed[uids] = FALSE
return(py_none())
},
# Carry out state changes associated with infection
set_prognoses = function(self, uids, sources = py_none()) {
ss$SIR$set_prognoses(self, uids, sources) # Perform SIR updates
# Define handling of exposure
ti <- self$ti # Current module time index
self$susceptible[uids] = FALSE # People exposed are no longer susceptible
self$exposed[uids] = TRUE # But they are exposed
self$ti_exposed[uids] = ti # Track what time they got infected
# Calculate and schedule future outcomes
dur_exp <- self$pars['dur_exp']$rvs(uids) # Get random numbers for the duration of exposure
self$ti_infected[uids] <- ti + dur_exp # Set the time at which people move from exposed to infected
dur_inf <- self$pars['dur_inf']$rvs(uids) # Set the duration of infection
will_die <- self$pars['p_death']$rvs(uids) # Choose who will die
self$ti_recovered[uids[!will_die]] <- ti + dur_inf[!will_die] # Schedule people to recover
self$ti_dead[uids[will_die]] <- ti + dur_inf[will_die] # Schedule people to die
# Update result count of new infections
self$results['new_infections'][self$ti] = length(uids) # Store the number of new infections in the results
return(py_none())
},
# Plot results using ggplot
plot = function(self) {
library(ggplot2)
df <- self$results$to_df() # Export results to an R data.frame
fig <- ggplot(df, aes(timevec)) +
geom_line(aes(y = n_susceptible, colour = "Susceptible")) +
geom_line(aes(y = n_exposed, colour = "Exposed")) +
geom_line(aes(y = n_infected, colour = "Infected")) +
geom_line(aes(y = n_recovered, colour = "Recovered"))
print(fig)
return(fig)
}
)
)
# Run and plot
seir <- SEIR() # Create the disease module
sim <- ss$Sim(diseases=seir, networks='random') # Create a simple sim with default parameters
sim$run() # Run
sim$diseases$seir$plot() # Call the custom ggplot function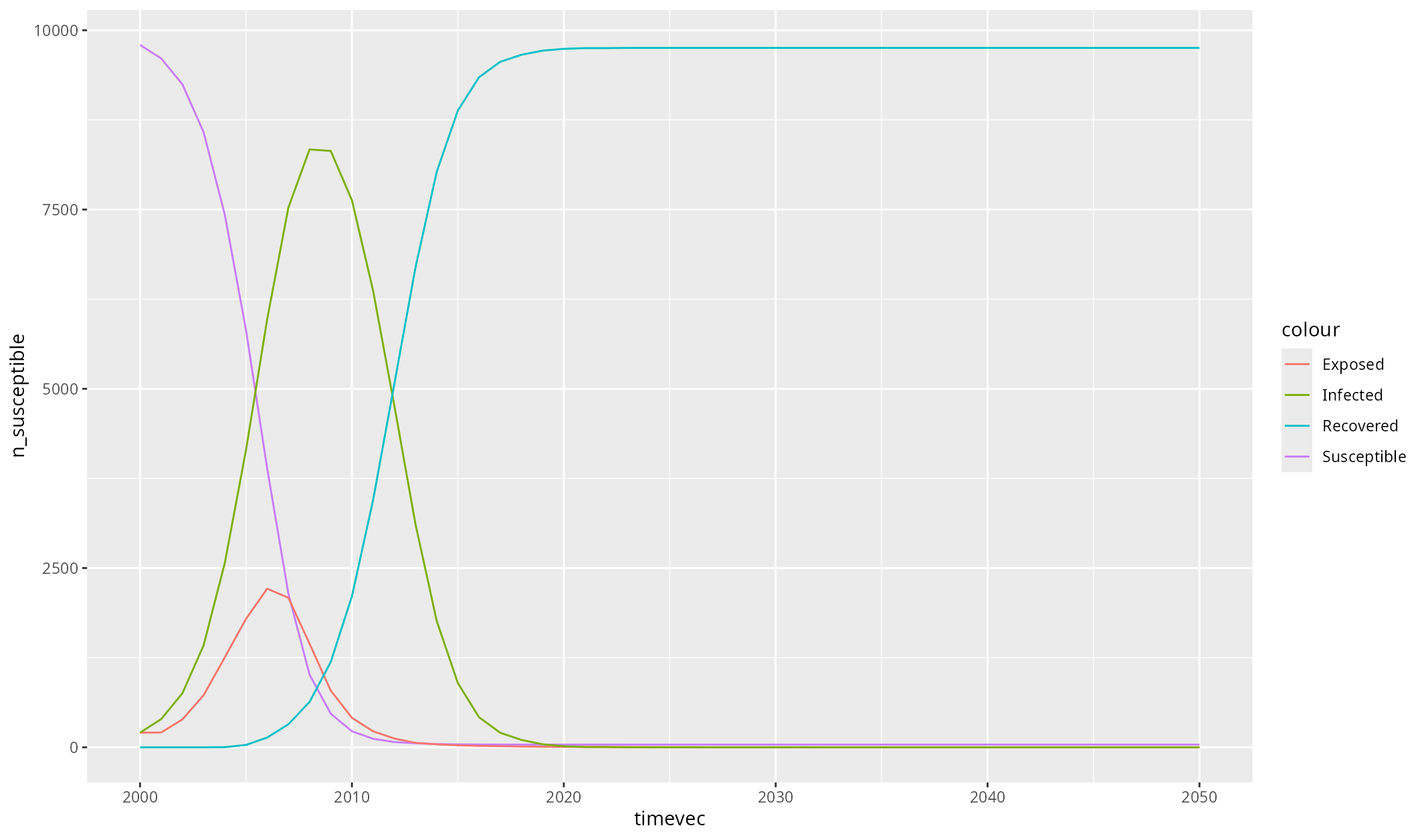
Next steps
For more intermediate examples between the simple example (which is very simple) and this one (which is very advanced), please see the Starsim documentation. If you still have questions, please reach out to us – we’re very happy to help!