Starsim is a framework for modeling the spread of diseases among agents via dynamic transmission networks. R-Starsim is a wrapper that facilitates the usage of Starsim from R. It uses reticulate to communicate between Python and R. Full documentation is available at https://r.starsim.org.
Installation
R-Starsim will be released on CRAN in future, but for now it can be installed via GitHub:
# install.packages("devtools")
devtools::install_github("starsimhub/rstarsim")
library(starsim)
init_starsim()If you do not already have a reticulate Python environment set up, R-Starsim will try to make one for you (by downloading Miniconda, creating an r-starsim environment, and activating it).
If you want to reinstall Starsim (e.g. to update the version), you can use:
If you already have Starsim installed in a Python (specifically conda) environment, you can skip installation and proceed directly to using Starsim in R. If the environment you’ve installed Starsim into is called starsim_env, then you can start using Starsim with:
library(starsim)
load_starsim("starsim_env")If you’ve installed it into your base conda environment, then you can simply use load_starsim("base").
Usage
All Starsim Python functions and classes are available in R. In Python, usage is typically import starsim as ss. To emulate this behavior in R, Starsim is made available as the variable ss, e.g. sim = ss.Sim() in Python becomes sim <- ss$Sim() in R. In addition, major Starsim classes (such as Sim, Network, Disease, etc.) are imported directly into the R namespace (e.g. sim <- Sim() also works).
Example
# Load Starsim
library(starsim)
load_starsim()
# Set the simulation parameters
pars <- list(
n_agents = 10000,
birth_rate = 20,
death_rate = 15,
networks = list(
type = 'randomnet',
n_contacts = 4
),
diseases = list(
type = 'sir',
dur_inf = 10,
beta = 0.1
)
)
# Create, run, and plot the simulation
sim <- ss$Sim(pars)
sim$run()
sim$diseases$sir$plot()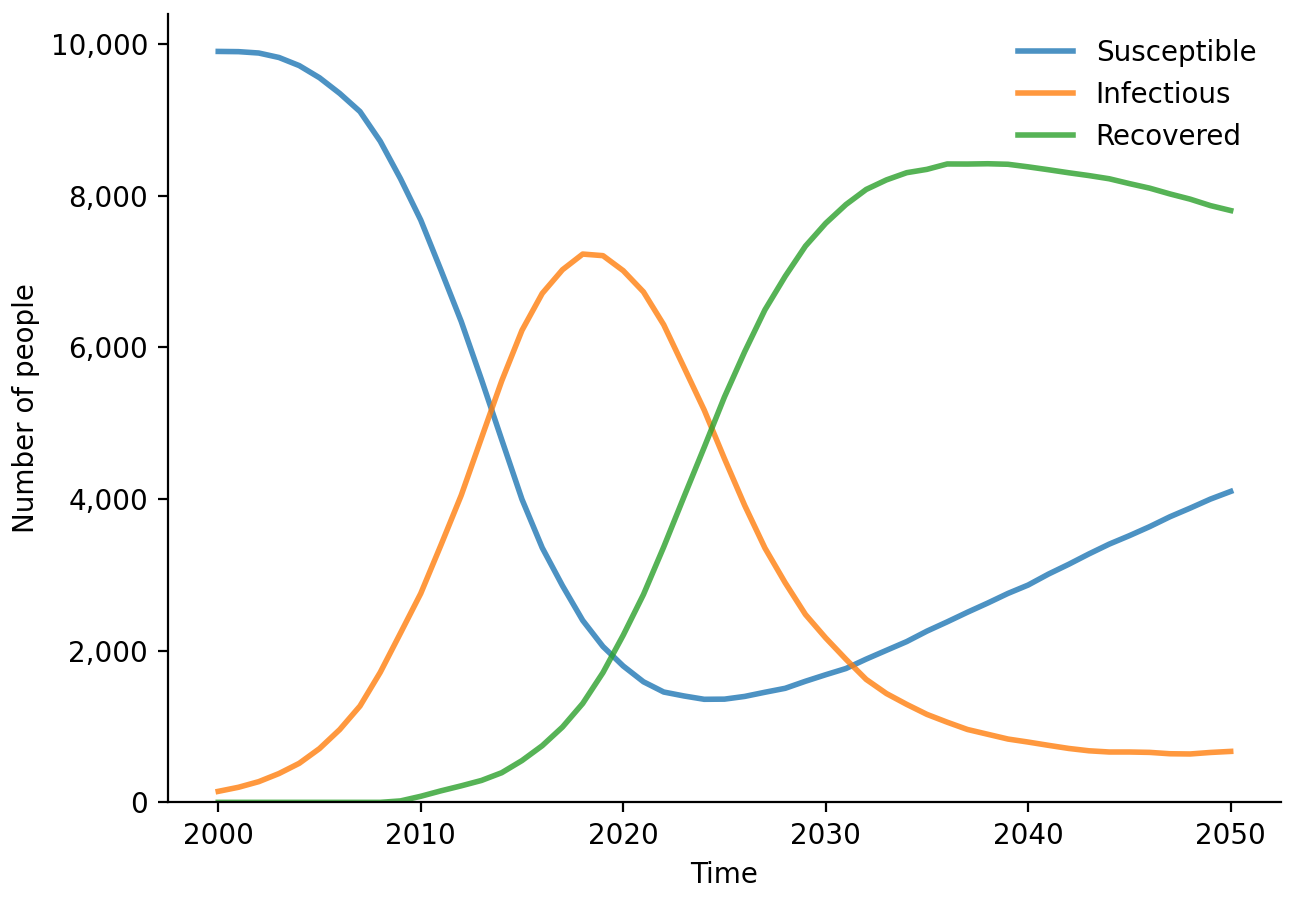
Contact
Questions? Comments? Bugs? Please open an issue or email us for help.